>>Our goals
1. Developing novel experimental and computational methods to discover new classes of RNAs and their functional mechanisms.
We have published papers in Nature Biotechnology, Nature Communications, Accounts of Chemical Research, etc.
We have developed RIP-PEN-seq, RIP-PEN-SHAPE-MaP, PEN-seq, NAP-seq and NAP-SHAPE-MaP sequencing technologies and developed computational software such as kturnSeeker and napSeeker.
2. Developing new experimental and computational technologies to identify RNA modifications and their functional mechanisms.
We have published papers in Nature, Nature Cell Biology, Sci China Life Sci, etc.
We have developed Nm-REP-seq sequencing technology and computational tools and platforms such as snoSeeker, RMBase and endSeeker.
3. Elucidating the regulatory networks and functional mechanisms of ncRNAs and their interacting RNA-binding proteins.
Published papers in Genome Research, Nucleic Acids Res,Cell Reports, etc.
We have developed new functional screening platforms and computational platforms such as starBase, ChIPBase and deepBase.
Recent Publications
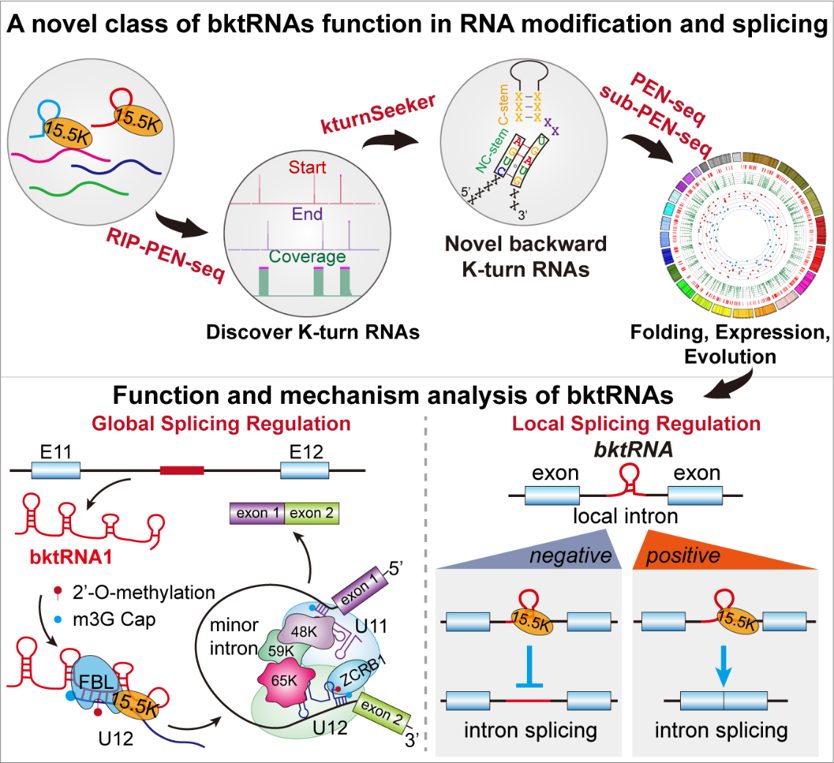
RIP-PEN-seq identifies a class of kink-turn RNAs as splicing regulators. Nature Biotechnology. 2024
A kink-turn (K-turn) is a three-dimensional RNA structure that exists in all three primary phylogenetic domains. In this study, we developed the RIP-PEN-seq method to identify the full-length sequences of RNAs bound by the K-turn binding protein 15.5K and discovered a previously uncharacterized class of RNAs with backward K-turn motifs (bktRNAs) in humans and mice. All bktRNAs share two consensus sequence motifs at their fixed terminal position and have complex folding properties, expression and evolution patterns. We found that a highly conserved bktRNA1 guides the methyltransferase fibrillarin to install RNA methylation of U12 small nuclear RNA in humans. Depletion of bktRNA1 causes global splicing dysregulation of U12-type introns by impairing the recruitment of ZCRB1 to the minor spliceosome. Most bktRNAs regulate the splicing of local introns by interacting with the 15.5K protein. Taken together, our findings characterize a class of small RNAs and uncover another layer of gene expression regulation that involves crosstalk among bktRNAs, RNA splicing and RNA methylation.
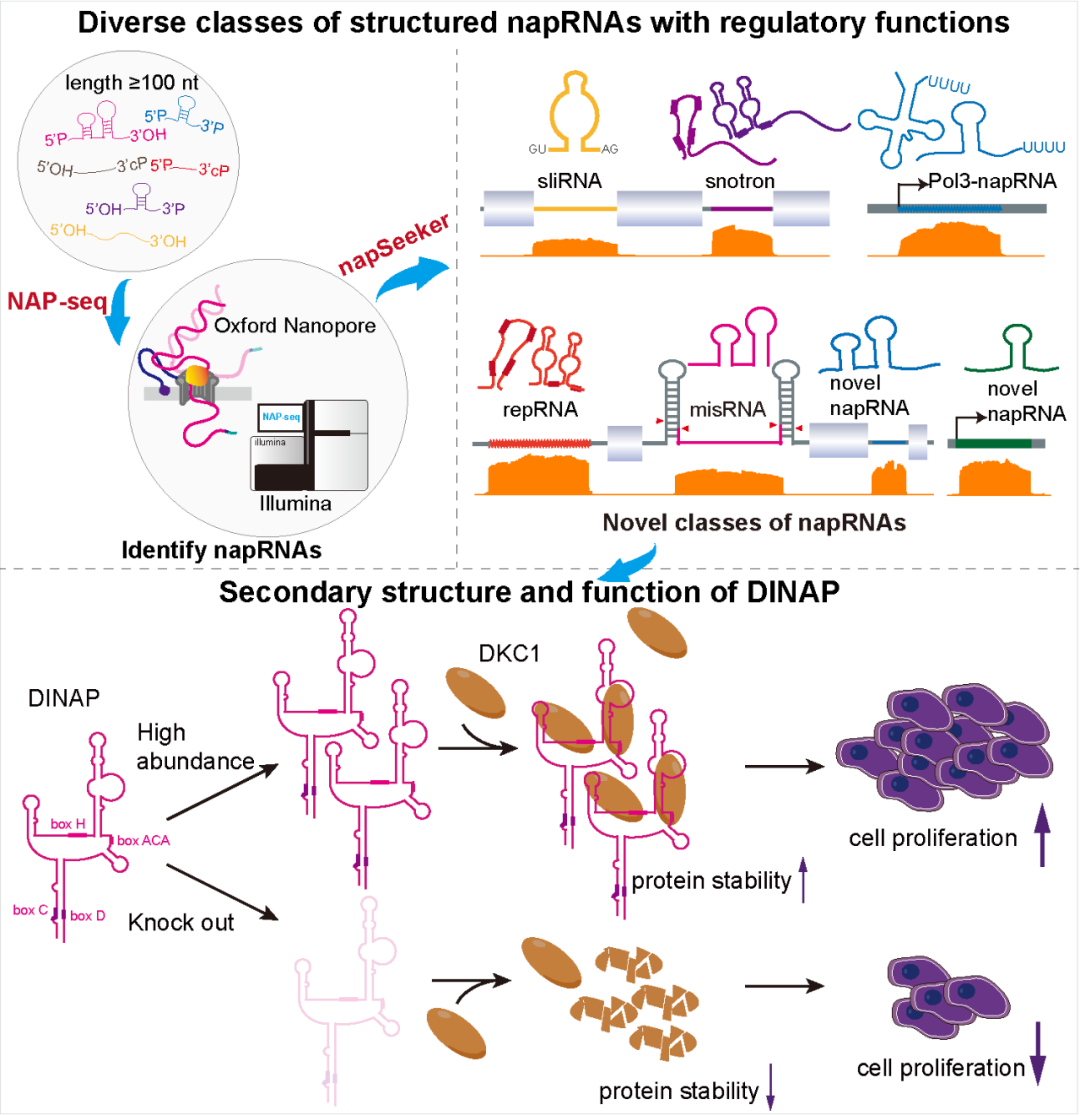
NAP-seq reveals multiple classes of structured noncoding RNAs with regulatory functions. Nature Communications. 2024
Up to 80% of the human genome produces “dark matter” RNAs, most of which are noncapped RNAs (napRNAs) that frequently act as noncoding RNAs (ncRNAs) to modulate gene expression.
Here, by developing a method, NAP-seq, to globally profile the full-length sequences of napRNAs with various terminal modifications at single-nucleotide resolution, we reveal diverse classes of structured ncRNAs. We discover stably expressed linear intron RNAs (sliRNAs), a class of snoRNA-intron RNAs (snotrons), a class of RNAs embedded in miRNA spacers (misRNAs) and thousands of previously uncharacterized structured napRNAs in humans and mice.
These napRNAs undergo dynamic changes in response to various stimuli and differentiation stages. Importantly, we show that a structured napRNA regulates myoblast differentiation and a napRNA DINAP interacts with dyskerin pseudouridine synthase 1 (DKC1) to promote cell proliferation by maintaining DKC1 protein stability. Our approach establishes a paradigm for discovering various classes of ncRNAs with regulatory functions.
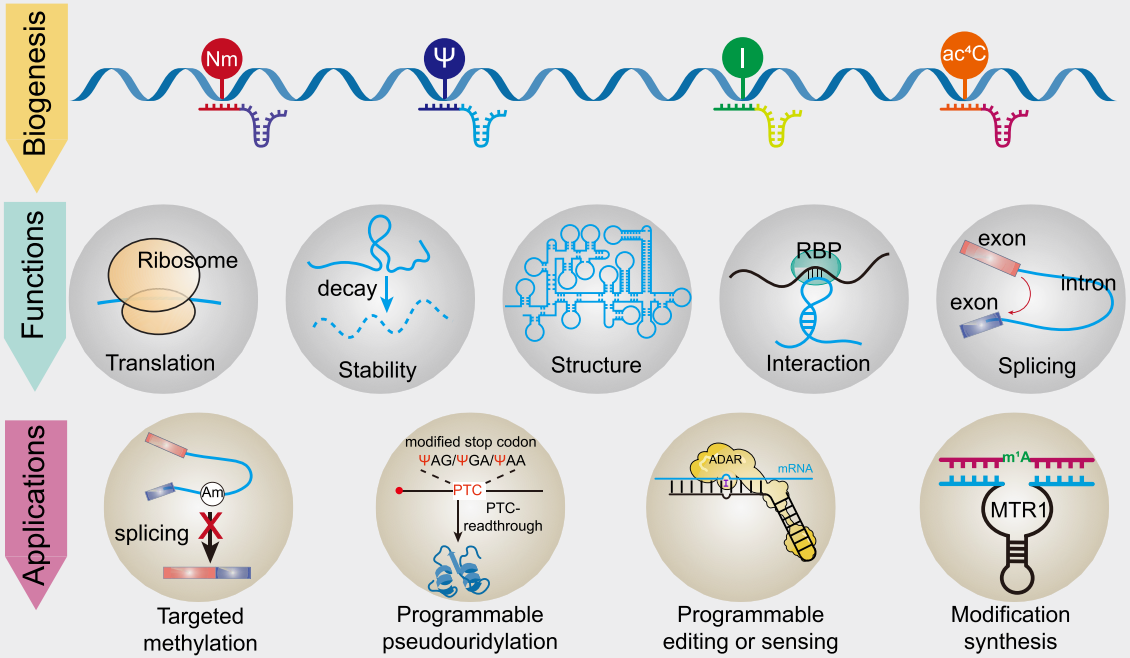
RNA-Guided RNA Modifications: Biogenesis, Functions, and Applications. Accounts of Chemical Research. 2023
In this Account, we focus on RNA modifications synthesized in an RNA-guided manner, including 2′-O-methylated nucleotides (Nm), pseudouridine (Ψ), N4-acetylcytidine (ac4C), and inosine (I). This Account sheds light on the intricate processes of biogenesis and elucidates the regulatory roles of these modifications in RNA metabolism. These roles include pivotal functions such as RNA stability, translation, and splicing, where each modification contributes to the diverse and finely tuned regulatory landscape of RNA biology.