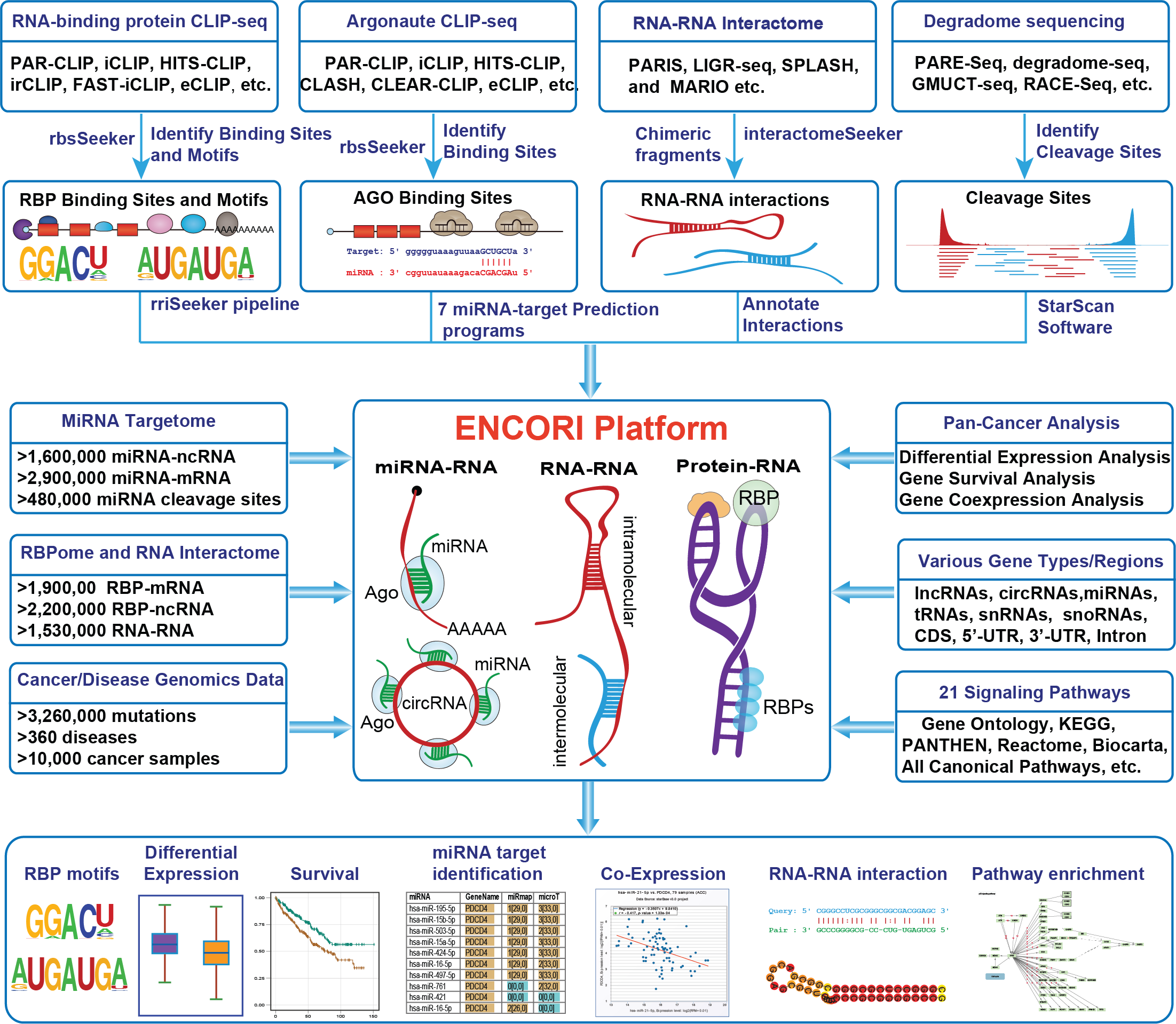
ENCORI/starBase (Encyclopedia of RNA Interactomes) is an extensive atlas that integrates precise RNA interactions identified by our innovative rbsSeeker and rriScan algorithms, showcasing the functional and mechanistic insights into the RNA interactomes.
Utilizing our new rbsSeeker software, we systematically identified distinct RNA-binding preferences for hundreds of RBPs (RNA-binding proteins), uncovering millions of high-confidence RBP-RNA interactions with diverse cellular functions and significant clinical implications in cancers. Additionally, our novel rriScan software revealed the complexity of RNA regulation by discovering millions of reliable RNA-RNA interactions.
Accompany with gene expression data of 32 types of cancers, ENCORI allows researchers performing Pan-Cancer analysis on RNA-RNA and RBP-RNA interactions. ENCORI also provides platforms to perform the survival and differential expression analysis of miRNAs, lncRNAs, pseudogenes and mRNAs.
How to cite:
- Zhou KR, Huang JH, Liu SR, Zheng WJ, Liu S, et al. An Encyclopedic Regulatory and Functional Atlas of RNA Interactomes. (Manuscript submitted)
- Li JH, et al.starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data , Nucleic Acids Res. 2014 Jan;42:D92-7.
ENCORI/starBase Citations (2015-2025) Cite Our Paper
>31,000 Citations in Google Scholar (starBase, the pilot version)- Average Daily Usage: ~5000 visits from ~500 unique researchers
- Average Annual Usage: >1,000,000 visits from >100 countries
Statistics of ENCORI (last update: 10/18/2024)
- Species: 23 Species
- CLIP-seq data: 2,725 datasets
- Degradome-seq: 100 datasets
- RNA-RNA interactome: 59 datasets
- RNA-seq data: >10,800 samples from 32 cancer types
- miRNA-seq data: >10,500 samples from 32 cancer types
- Disease data: >1,800,000 mutations from 531 disease types
- miRNA-ncRNA(CLIP): >460,000 interactions
- miRNA-mRNA(CLIP): >1,200,000 interactions
- RBP-mRNA: >1,290,000 interactions
- RBP-ncRNA: >1,600,000 interactions
- RNA-RNA: >3,700,000 interactions
- miRNA-ncRNA(degradome): >32,000 interactions
- miRNA-mRNA(degradome): >459,000 interactions
- ceRNA: >2,900,000 pairs
- function annoation: >34,000 functional terms from 21 categories
- Pan-Cancer: Differential Expression,Survival Analysis,CoExpression
For better browse experience, Chrome is strongly recommended. Firefox 61, Opera 54, Safari 5 and Microsoft Edge are also supported.
IE 7-11 and IE-core browsers are not supported.